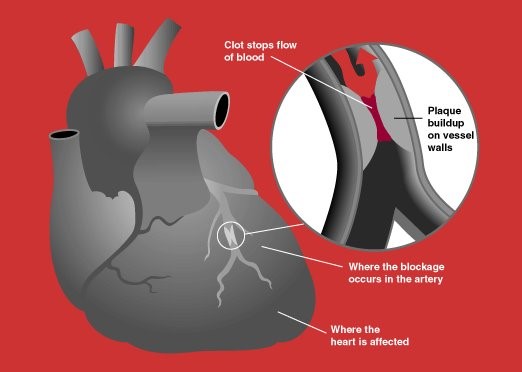
Photo by Creative Commons Attribution Share-Alike (CCYYSA)
 Loading…
Loading…
Exploratory Data Analysis
In [1]:
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
In [2]:
df = pd.read_csv('heart_dataset.csv')
In [3]:
df.head()
Out[3]:
| age | sex | cp | trestbps | chol | fbs | restecg | thalachh | exang | oldpeak | slope | ca | thal | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 63 | 1 | 3 | 145 | 233 | 1 | 0 | 150 | 0 | 2.3 | 0 | 0 | 1 | 1 |
| 1 | 37 | 1 | 2 | 130 | 250 | 0 | 1 | 187 | 0 | 3.5 | 0 | 0 | 2 | 1 |
| 2 | 41 | 0 | 1 | 130 | 204 | 0 | 0 | 172 | 0 | 1.4 | 2 | 0 | 2 | 1 |
| 3 | 56 | 1 | 1 | 120 | 236 | 0 | 1 | 178 | 0 | 0.8 | 2 | 0 | 2 | 1 |
| 4 | 57 | 0 | 0 | 120 | 354 | 0 | 1 | 163 | 1 | 0.6 | 2 | 0 | 2 | 1 |
In [4]:
df.info()
<class 'pandas.core.frame.DataFrame'> RangeIndex: 1888 entries, 0 to 1887 Data columns (total 14 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 age 1888 non-null int64 1 sex 1888 non-null int64 2 cp 1888 non-null int64 3 trestbps 1888 non-null int64 4 chol 1888 non-null int64 5 fbs 1888 non-null int64 6 restecg 1888 non-null int64 7 thalachh 1888 non-null int64 8 exang 1888 non-null int64 9 oldpeak 1888 non-null float64 10 slope 1888 non-null int64 11 ca 1888 non-null int64 12 thal 1888 non-null int64 13 target 1888 non-null int64 dtypes: float64(1), int64(13) memory usage: 206.6 KB
In [5]:
df.describe()
Out[5]:
| age | sex | cp | trestbps | chol | fbs | restecg | thalachh | exang | oldpeak | slope | ca | thal | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 1888.000000 | 1888.000000 | 1888.000000 | 1888.000000 | 1888.000000 | 1888.000000 | 1888.000000 | 1888.000000 | 1888.000000 | 1888.000000 | 1888.000000 | 1888.000000 | 1888.000000 | 1888.000000 |
| mean | 54.354343 | 0.688559 | 1.279131 | 131.549258 | 246.855403 | 0.148305 | 0.597458 | 149.424258 | 0.331568 | 1.053761 | 1.421610 | 0.731462 | 2.662606 | 0.517479 |
| std | 9.081505 | 0.463205 | 1.280877 | 17.556985 | 51.609329 | 0.355496 | 0.638820 | 23.006153 | 0.470901 | 1.161344 | 0.619588 | 1.015735 | 1.249924 | 0.499827 |
| min | 29.000000 | 0.000000 | 0.000000 | 94.000000 | 126.000000 | 0.000000 | 0.000000 | 71.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 47.750000 | 0.000000 | 0.000000 | 120.000000 | 211.000000 | 0.000000 | 0.000000 | 133.000000 | 0.000000 | 0.000000 | 1.000000 | 0.000000 | 2.000000 | 0.000000 |
| 50% | 55.000000 | 1.000000 | 1.000000 | 130.000000 | 241.000000 | 0.000000 | 1.000000 | 152.000000 | 0.000000 | 0.800000 | 1.000000 | 0.000000 | 2.000000 | 1.000000 |
| 75% | 61.000000 | 1.000000 | 2.000000 | 140.000000 | 276.000000 | 0.000000 | 1.000000 | 166.000000 | 1.000000 | 1.600000 | 2.000000 | 1.000000 | 3.000000 | 1.000000 |
| max | 77.000000 | 1.000000 | 4.000000 | 200.000000 | 564.000000 | 1.000000 | 2.000000 | 202.000000 | 1.000000 | 6.200000 | 3.000000 | 4.000000 | 7.000000 | 1.000000 |
In [6]:
df.describe().T
Out[6]:
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| age | 1888.0 | 54.354343 | 9.081505 | 29.0 | 47.75 | 55.0 | 61.0 | 77.0 |
| sex | 1888.0 | 0.688559 | 0.463205 | 0.0 | 0.00 | 1.0 | 1.0 | 1.0 |
| cp | 1888.0 | 1.279131 | 1.280877 | 0.0 | 0.00 | 1.0 | 2.0 | 4.0 |
| trestbps | 1888.0 | 131.549258 | 17.556985 | 94.0 | 120.00 | 130.0 | 140.0 | 200.0 |
| chol | 1888.0 | 246.855403 | 51.609329 | 126.0 | 211.00 | 241.0 | 276.0 | 564.0 |
| fbs | 1888.0 | 0.148305 | 0.355496 | 0.0 | 0.00 | 0.0 | 0.0 | 1.0 |
| restecg | 1888.0 | 0.597458 | 0.638820 | 0.0 | 0.00 | 1.0 | 1.0 | 2.0 |
| thalachh | 1888.0 | 149.424258 | 23.006153 | 71.0 | 133.00 | 152.0 | 166.0 | 202.0 |
| exang | 1888.0 | 0.331568 | 0.470901 | 0.0 | 0.00 | 0.0 | 1.0 | 1.0 |
| oldpeak | 1888.0 | 1.053761 | 1.161344 | 0.0 | 0.00 | 0.8 | 1.6 | 6.2 |
| slope | 1888.0 | 1.421610 | 0.619588 | 0.0 | 1.00 | 1.0 | 2.0 | 3.0 |
| ca | 1888.0 | 0.731462 | 1.015735 | 0.0 | 0.00 | 0.0 | 1.0 | 4.0 |
| thal | 1888.0 | 2.662606 | 1.249924 | 0.0 | 2.00 | 2.0 | 3.0 | 7.0 |
| target | 1888.0 | 0.517479 | 0.499827 | 0.0 | 0.00 | 1.0 | 1.0 | 1.0 |
In [7]:
# List of numeric variables
numeric_vars = ['age', 'trestbps', 'chol', 'thalachh', 'oldpeak']
# Create distplots for each numeric variable
for var in numeric_vars:
plt.figure(figsize=(10, 6))
sns.histplot(df[var], kde=True, bins=30)
plt.title(f'Distribution of {var}')
plt.xlabel(var)
plt.ylabel('Density')
plt.show()
In [8]:
categorical_vars = ['sex', 'cp', 'fbs', 'restecg', 'exang', 'slope', 'ca', 'thal', 'target']
for var in categorical_vars:
plt.figure(figsize=(8, 6))
df[var].value_counts().plot.pie(autopct='%1.1f%%', startangle=90, cmap='Pastel1')
plt.title(f'Distribution of {var}')
plt.ylabel('')
plt.show()
In [9]:
from sklearn.preprocessing import StandardScaler
# Initialize the scaler
scaler = StandardScaler()
# Fit and transform the numeric variables
df[numeric_vars] = scaler.fit_transform(df[numeric_vars])
# Display the first few rows of the scaled dataframe
df.head()
Out[9]:
| age | sex | cp | trestbps | chol | fbs | restecg | thalachh | exang | oldpeak | slope | ca | thal | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.952259 | 1 | 3 | 0.766322 | -0.268538 | 1 | 0 | 0.025032 | 0 | 1.073386 | 0 | 0 | 1 | 1 |
| 1 | -1.911461 | 1 | 2 | -0.088265 | 0.060947 | 0 | 1 | 1.633724 | 0 | 2.106945 | 0 | 0 | 2 | 1 |
| 2 | -1.470888 | 0 | 1 | -0.088265 | -0.830601 | 0 | 0 | 0.981551 | 0 | 0.298216 | 2 | 0 | 2 | 1 |
| 3 | 0.181258 | 1 | 1 | -0.657990 | -0.210394 | 0 | 1 | 1.242420 | 0 | -0.218564 | 2 | 0 | 2 | 1 |
| 4 | 0.291401 | 0 | 0 | -0.657990 | 2.076620 | 0 | 1 | 0.590248 | 1 | -0.390824 | 2 | 0 | 2 | 1 |
In [10]:
# Calculate the correlation matrix
corr_matrix = df.corr()
# Create a heatmap
plt.figure(figsize=(12, 8))
sns.heatmap(corr_matrix, annot=True, fmt='.2f', cmap='coolwarm', linewidths=0.5)
plt.title('Correlation Heatmap')
plt.show()
In [ ]:
Logistic Regression
In [1]:
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
In [2]:
df = pd.read_csv('C:\\Users\\Dell\\Desktop\\Python Notebooks\\AI 201\\HEART ATTACK\\heart_dataset.csv')
In [3]:
X = df.iloc[:, :-1].values
y = df.iloc[:, -1].values
In [4]:
# One Hot Encoding columns Sex, CP, FBS, RestECG, Exang, Slope, CA, and Thal
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import OneHotEncoder
ct = ColumnTransformer(transformers=[('encoder', OneHotEncoder(), [1, 2, 5, 6, 8, 10, 11, 12])], remainder='passthrough')
X = np.array(ct.fit_transform(X))
In [5]:
# Feature Scaling
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X[:, [0, 3, 4, 7, 9]] = sc.fit_transform(X[:, [0, 3, 4, 7, 9]])
In [6]:
# Splitting the dataset into the Training set and Test set
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state = 1)
In [7]:
# Training the Logistic Regression model on the Training set
from sklearn.linear_model import LogisticRegression
classifier = LogisticRegression(C=0.1, max_iter=300, penalty='l2', solver='lbfgs', random_state = 0)
classifier.fit(X_train, y_train)
c:\Users\Dell\anaconda3\Lib\site-packages\sklearn\linear_model\_logistic.py:469: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
Please also refer to the documentation for alternative solver options:
https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression
n_iter_i = _check_optimize_result(
Out[7]:
LogisticRegression(C=0.1, max_iter=300, random_state=0)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
LogisticRegression(C=0.1, max_iter=300, random_state=0)
In [8]:
# Predicting the Test set results
y_pred = classifier.predict(X_test)
In [9]:
# Making the Confusion Matrix
from sklearn.metrics import confusion_matrix, accuracy_score, precision_score, recall_score, f1_score
cm = confusion_matrix(y_test, y_pred)
print(cm)
print(accuracy_score(y_test, y_pred))
[[139 31] [ 36 172]] 0.8227513227513228
In [10]:
from sklearn.model_selection import cross_val_score
scores = cross_val_score(classifier, X_test, y_test, cv = 10)
print("Cross-Validation Accuracy Scores", scores.mean())
c:\Users\Dell\anaconda3\Lib\site-packages\sklearn\linear_model\_logistic.py:469: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
Please also refer to the documentation for alternative solver options:
https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression
n_iter_i = _check_optimize_result(
c:\Users\Dell\anaconda3\Lib\site-packages\sklearn\linear_model\_logistic.py:469: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
Please also refer to the documentation for alternative solver options:
https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression
n_iter_i = _check_optimize_result(
c:\Users\Dell\anaconda3\Lib\site-packages\sklearn\linear_model\_logistic.py:469: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
Please also refer to the documentation for alternative solver options:
https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression
n_iter_i = _check_optimize_result(
c:\Users\Dell\anaconda3\Lib\site-packages\sklearn\linear_model\_logistic.py:469: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
Please also refer to the documentation for alternative solver options:
https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression
n_iter_i = _check_optimize_result(
c:\Users\Dell\anaconda3\Lib\site-packages\sklearn\linear_model\_logistic.py:469: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
Please also refer to the documentation for alternative solver options:
https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression
n_iter_i = _check_optimize_result(
c:\Users\Dell\anaconda3\Lib\site-packages\sklearn\linear_model\_logistic.py:469: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
Please also refer to the documentation for alternative solver options:
https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression
n_iter_i = _check_optimize_result(
c:\Users\Dell\anaconda3\Lib\site-packages\sklearn\linear_model\_logistic.py:469: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
Please also refer to the documentation for alternative solver options:
https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression
n_iter_i = _check_optimize_result(
c:\Users\Dell\anaconda3\Lib\site-packages\sklearn\linear_model\_logistic.py:469: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
Please also refer to the documentation for alternative solver options:
https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression
n_iter_i = _check_optimize_result(
Cross-Validation Accuracy Scores 0.8043385490753913
c:\Users\Dell\anaconda3\Lib\site-packages\sklearn\linear_model\_logistic.py:469: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
Please also refer to the documentation for alternative solver options:
https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression
n_iter_i = _check_optimize_result(
c:\Users\Dell\anaconda3\Lib\site-packages\sklearn\linear_model\_logistic.py:469: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
Please also refer to the documentation for alternative solver options:
https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression
n_iter_i = _check_optimize_result(
In [11]:
accuracy = accuracy_score(y_test, y_pred)
precision = precision_score(y_test, y_pred, average='weighted')
recall = recall_score(y_test, y_pred, average='weighted')
f1 = f1_score(y_test, y_pred, average='weighted')
print("Accuracy Score:", accuracy)
print("Precision Score:", precision)
print("Recall Score:", recall)
print("F1 Score:", f1)
Accuracy Score: 0.8227513227513228 Precision Score: 0.8234524461125446 Recall Score: 0.8227513227513228 F1 Score: 0.8229575784849243
In [12]:
#plot_roc_curve(classifier, X_test, y_test, name = "Logistic Regression")
#plt.title("Logistic Regression Roc Curve And AUC")
#plt.plot([0, 1], [0, 1], "r--")
#plt.show()
from sklearn.metrics import RocCurveDisplay
RocCurveDisplay.from_estimator(estimator=classifier, X=X_test, y=y_test, name = "Logistic Regression")
plt.title("Logistic Regression Roc Curve And AUC")
plt.plot([0, 1], [0, 1], "r--")
plt.show()
In [13]:
# Define the parameter grid
# param_grid = {
# 'penalty': ['l1', 'l2', 'elasticnet', 'none'],
# 'C': [0.01, 0.1, 1, 10, 100],
# 'solver': ['newton-cg', 'lbfgs', 'liblinear', 'sag', 'saga'],
# 'max_iter': [100, 200, 300]
# }
# # Initialize the GridSearchCV object
# grid_search = GridSearchCV(estimator=classifier, param_grid=param_grid, cv=10, n_jobs=-1, scoring='accuracy')
# # Fit the grid search to the data
# grid_search.fit(X_train, y_train)
# # Print the best parameters and best score
# print("Best Parameters:", grid_search.best_params_)
# print("Best Score:", grid_search.best_score_)
Naive Bayes
In [1]:
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import GridSearchCV
In [2]:
df = pd.read_csv('C:\\Users\\Dell\\Desktop\\Python Notebooks\\AI 201\\HEART ATTACK\\heart_dataset.csv')
In [3]:
X = df.iloc[:, :-1].values
y = df.iloc[:, -1].values
In [4]:
# One Hot Encoding columns Sex, CP, FBS, RestECG, Exang, Slope, CA, and Thal
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import OneHotEncoder
ct = ColumnTransformer(transformers=[('encoder', OneHotEncoder(), [1, 2, 5, 6, 8, 10, 11, 12])], remainder='passthrough')
X = np.array(ct.fit_transform(X))
In [5]:
# Feature Scaling
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X[:, [0, 3, 4, 7, 9]] = sc.fit_transform(X[:, [0, 3, 4, 7, 9]])
In [6]:
# Splitting the dataset into the Training set and Test set
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state = 1)
In [7]:
# Training the Naive Bayes model on the Training set
from sklearn.naive_bayes import GaussianNB
classifier = GaussianNB()
classifier.fit(X_train, y_train)
Out[7]:
GaussianNB()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
GaussianNB()
In [8]:
# Predicting the Test set results
y_pred = classifier.predict(X_test)
In [9]:
# Making the Confusion Matrix
from sklearn.metrics import confusion_matrix, accuracy_score, precision_score, recall_score, f1_score
cm = confusion_matrix(y_test, y_pred)
print(cm)
print(accuracy_score(y_test, y_pred))
[[134 36] [ 39 169]] 0.8015873015873016
In [10]:
from sklearn.model_selection import cross_val_score
scores = cross_val_score(classifier, X_test, y_test, cv = 10)
print("Cross-Validation Accuracy Scores", scores.mean())
Cross-Validation Accuracy Scores 0.7331436699857752
In [11]:
from sklearn.metrics import RocCurveDisplay
RocCurveDisplay.from_estimator(estimator=classifier, X=X_test, y=y_test, name = "Naive Bayes")
plt.title("Naive Bayes Roc Curve And AUC")
plt.plot([0, 1], [0, 1], "r--")
plt.show()
In [12]:
# Hyperparameter optimization using GridSearchCV
param_grid = {
'var_smoothing': np.logspace(0, -9, num=100)
}
grid_search = GridSearchCV(estimator=GaussianNB(), param_grid=param_grid, cv=10, n_jobs=-1, scoring='accuracy')
grid_search.fit(X_train, y_train)
print("Best parameters found: ", grid_search.best_params_)
print("Best cross-validation accuracy: ", grid_search.best_score_)
Best parameters found: {'var_smoothing': 3.511191734215127e-05}
Best cross-validation accuracy: 0.7867549668874172
In [13]:
# Evaluating the model with the best found parameters on the test set
best_classifier = grid_search.best_estimator_
y_pred_best = best_classifier.predict(X_test)
cm_best = confusion_matrix(y_test, y_pred_best)
print("Confusion Matrix with best parameters:\n", cm_best)
print("Accuracy Score with best parameters:", accuracy_score(y_test, y_pred_best))
accuracy = accuracy_score(y_test, y_pred_best)
precision = precision_score(y_test, y_pred_best, average='weighted')
recall = recall_score(y_test, y_pred_best, average='weighted')
f1 = f1_score(y_test, y_pred_best, average='weighted')
print("Accuracy Score:", accuracy)
print("Precision Score:", precision)
print("Recall Score:", recall)
print("F1 Score:", f1)
Confusion Matrix with best parameters: [[135 35] [ 44 164]] Accuracy Score with best parameters: 0.791005291005291 Accuracy Score: 0.791005291005291 Precision Score: 0.7926703066929338 Recall Score: 0.791005291005291 F1 Score: 0.7913893128792842
In [14]:
RocCurveDisplay.from_estimator(estimator=best_classifier, X=X_test, y=y_test, name="Naive Bayes (Best Params)")
plt.title("Naive Bayes Roc Curve And AUC (Best Params)")
plt.plot([0, 1], [0, 1], "r--")
plt.show()
In [ ]:
Artificial Neural Network
In [1]:
from google.colab import drive
drive.mount('/content/drive')
Mounted at /content/drive
In [2]:
import numpy as np
import pandas as pd
import tensorflow as tf
import matplotlib.pyplot as plt
from sklearn.metrics import confusion_matrix, accuracy_score, precision_score, recall_score, f1_score
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import OneHotEncoder
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.model_selection import cross_val_score, KFold
from sklearn.metrics import RocCurveDisplay
In [3]:
tf.__version__
Out[3]:
'2.17.1'
In [4]:
df = pd.read_csv('/content/drive/MyDrive/ARTIFICIAL INTELLIGENCE/AI 201/FOR IN PERSON PRESENTATION/heart_dataset.csv')
X = df.iloc[:, :-1].values
y = df.iloc[:, -1].values
In [5]:
# One Hot Encoding columns Sex, CP, FBS, RestECG, Exang, Slope, CA, and Thal
ct = ColumnTransformer(transformers=[('encoder', OneHotEncoder(), [1, 2, 5, 6, 8, 10, 11, 12])], remainder='passthrough')
X = np.array(ct.fit_transform(X))
# Feature Scaling
sc = StandardScaler()
X[:, [0, 3, 4, 7, 9]] = sc.fit_transform(X[:, [0, 3, 4, 7, 9]])
In [6]:
# Splitting the dataset into the Training set and Test set
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state = 1)
In [7]:
ann = tf.keras.models.Sequential()
In [8]:
ann.add(tf.keras.layers.Dense(units=6, activation='relu'))
In [9]:
ann.add(tf.keras.layers.Dense(units=6, activation='relu'))
In [10]:
ann.add(tf.keras.layers.Dense(units=1, activation='sigmoid'))
In [11]:
ann.compile(optimizer = 'adam', loss = 'binary_crossentropy', metrics = ['accuracy'])
In [12]:
ann.fit(X_train, y_train, batch_size = 32, epochs = 100)
Epoch 1/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 2s 2ms/step - accuracy: 0.6209 - loss: 1.2411 Epoch 2/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.6153 - loss: 0.8739 Epoch 3/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 7ms/step - accuracy: 0.6113 - loss: 0.8462 Epoch 4/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.6216 - loss: 0.7336 Epoch 5/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.6504 - loss: 0.6834 Epoch 6/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.6337 - loss: 0.6596 Epoch 7/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.6748 - loss: 0.6172 Epoch 8/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.6750 - loss: 0.6145 Epoch 9/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.6832 - loss: 0.5821 Epoch 10/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.6947 - loss: 0.5568 Epoch 11/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 3ms/step - accuracy: 0.6924 - loss: 0.5701 Epoch 12/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7099 - loss: 0.5461 Epoch 13/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.6984 - loss: 0.5537 Epoch 14/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7303 - loss: 0.5270 Epoch 15/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7132 - loss: 0.5483 Epoch 16/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7294 - loss: 0.5084 Epoch 17/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7217 - loss: 0.5254 Epoch 18/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 3ms/step - accuracy: 0.7294 - loss: 0.5156 Epoch 19/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7338 - loss: 0.5015 Epoch 20/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7471 - loss: 0.4994 Epoch 21/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7517 - loss: 0.4830 Epoch 22/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7636 - loss: 0.4783 Epoch 23/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7525 - loss: 0.4786 Epoch 24/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7669 - loss: 0.4657 Epoch 25/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 3ms/step - accuracy: 0.7541 - loss: 0.4791 Epoch 26/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7527 - loss: 0.4696 Epoch 27/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7595 - loss: 0.4655 Epoch 28/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7657 - loss: 0.4502 Epoch 29/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7706 - loss: 0.4515 Epoch 30/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 3ms/step - accuracy: 0.7798 - loss: 0.4472 Epoch 31/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7692 - loss: 0.4395 Epoch 32/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7809 - loss: 0.4392 Epoch 33/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7947 - loss: 0.4170 Epoch 34/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7656 - loss: 0.4564 Epoch 35/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 4ms/step - accuracy: 0.7669 - loss: 0.4506 Epoch 36/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 3ms/step - accuracy: 0.7785 - loss: 0.4283 Epoch 37/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 3ms/step - accuracy: 0.7858 - loss: 0.4273 Epoch 38/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 3ms/step - accuracy: 0.8189 - loss: 0.3949 Epoch 39/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 4ms/step - accuracy: 0.8016 - loss: 0.4074 Epoch 40/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 3ms/step - accuracy: 0.7897 - loss: 0.4219 Epoch 41/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 4ms/step - accuracy: 0.7937 - loss: 0.4241 Epoch 42/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 3ms/step - accuracy: 0.8030 - loss: 0.4093 Epoch 43/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 3ms/step - accuracy: 0.7840 - loss: 0.4192 Epoch 44/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 3ms/step - accuracy: 0.7985 - loss: 0.4379 Epoch 45/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 3ms/step - accuracy: 0.7960 - loss: 0.4128 Epoch 46/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 4ms/step - accuracy: 0.8079 - loss: 0.4224 Epoch 47/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 4ms/step - accuracy: 0.8155 - loss: 0.4016 Epoch 48/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 3ms/step - accuracy: 0.7996 - loss: 0.4060 Epoch 49/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7848 - loss: 0.4591 Epoch 50/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8106 - loss: 0.3994 Epoch 51/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8253 - loss: 0.3829 Epoch 52/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7994 - loss: 0.3947 Epoch 53/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8317 - loss: 0.3736 Epoch 54/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8131 - loss: 0.3968 Epoch 55/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7945 - loss: 0.4178 Epoch 56/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 3ms/step - accuracy: 0.8035 - loss: 0.4187 Epoch 57/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7862 - loss: 0.4444 Epoch 58/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8030 - loss: 0.4062 Epoch 59/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8093 - loss: 0.4157 Epoch 60/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8162 - loss: 0.3970 Epoch 61/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8081 - loss: 0.4241 Epoch 62/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8085 - loss: 0.4160 Epoch 63/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 3ms/step - accuracy: 0.7977 - loss: 0.4202 Epoch 64/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8094 - loss: 0.4067 Epoch 65/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8167 - loss: 0.3961 Epoch 66/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8282 - loss: 0.3934 Epoch 67/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8156 - loss: 0.4060 Epoch 68/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8182 - loss: 0.3985 Epoch 69/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8113 - loss: 0.4008 Epoch 70/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8002 - loss: 0.4059 Epoch 71/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8270 - loss: 0.3929 Epoch 72/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8152 - loss: 0.4018 Epoch 73/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8224 - loss: 0.3971 Epoch 74/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8252 - loss: 0.3878 Epoch 75/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8368 - loss: 0.3825 Epoch 76/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8306 - loss: 0.3762 Epoch 77/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8183 - loss: 0.4025 Epoch 78/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 3ms/step - accuracy: 0.8129 - loss: 0.3980 Epoch 79/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.7880 - loss: 0.4325 Epoch 80/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8309 - loss: 0.3850 Epoch 81/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8052 - loss: 0.4209 Epoch 82/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8391 - loss: 0.3665 Epoch 83/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8116 - loss: 0.4003 Epoch 84/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8177 - loss: 0.3865 Epoch 85/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8108 - loss: 0.4145 Epoch 86/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8285 - loss: 0.3924 Epoch 87/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8349 - loss: 0.3936 Epoch 88/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8101 - loss: 0.4180 Epoch 89/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8158 - loss: 0.4174 Epoch 90/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8245 - loss: 0.3724 Epoch 91/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 3ms/step - accuracy: 0.8407 - loss: 0.3704 Epoch 92/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8245 - loss: 0.3955 Epoch 93/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8138 - loss: 0.4078 Epoch 94/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8299 - loss: 0.3807 Epoch 95/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8018 - loss: 0.4280 Epoch 96/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8166 - loss: 0.4264 Epoch 97/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8173 - loss: 0.3928 Epoch 98/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8227 - loss: 0.4052 Epoch 99/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8489 - loss: 0.3765 Epoch 100/100 48/48 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step - accuracy: 0.8214 - loss: 0.3850
Out[12]:
<keras.src.callbacks.history.History at 0x7d9eafaa9780>
In [13]:
y_pred = ann.predict(X_test)
y_pred = (y_pred > 0.5)
12/12 ━━━━━━━━━━━━━━━━━━━━ 0s 5ms/step
In [14]:
from sklearn.metrics import confusion_matrix, accuracy_score
cm = confusion_matrix(y_test, y_pred)
print(cm)
print("Accuracy Score", accuracy_score(y_test, y_pred))
[[132 38] [ 27 181]] Accuracy Score 0.828042328042328
In [15]:
accuracy = accuracy_score(y_test, y_pred)
precision = precision_score(y_test, y_pred, average='weighted')
recall = recall_score(y_test, y_pred, average='weighted')
f1 = f1_score(y_test, y_pred, average='weighted')
print("Accuracy Score:", accuracy)
print("Precision Score:", precision)
print("Recall Score:", recall)
print("F1 Score:", f1)
Accuracy Score: 0.828042328042328 Precision Score: 0.828150135636997 Recall Score: 0.828042328042328 F1 Score: 0.8273825671802651
In [16]:
from sklearn.metrics import roc_curve, auc
In [17]:
# Plotting the ROC Curve
# Compute predicted probabilities
y_pred_prob = ann.predict(X_test).ravel()
# Compute ROC curve and ROC area
fpr, tpr, thresholds = roc_curve(y_test, y_pred_prob)
roc_auc = auc(fpr, tpr)
# Plot ROC curve
plt.figure()
plt.plot(fpr, tpr, color='darkorange', lw=2, label='ROC curve (area = %0.2f)' % roc_auc)
plt.plot([0, 1], [0, 1], color='navy', lw=2, linestyle='--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('ANN ROC Curve and AUC')
plt.legend(loc="lower right")
plt.show()
12/12 ━━━━━━━━━━━━━━━━━━━━ 0s 2ms/step